2. The dolphin slaughter took place in Taiji Japan
4. Ric O-Barry's turning point that completely changed and "turn his Porches in for this crusade" is when he experienced the suicide of Flipper, his dolphin, because she was suffering from being held captive.
6. The country that indirectly runs the International Whaling Commission is Japan
8. Fishermen trap the dolphins in the cove by banging metal rods which scares the dolphins away from the noise and towards the cove. Then the cove is blocked so that thousands of dolphins are captured.
10. 23,000 every year.
12. The toxic substance is mercury.
14. Japanese people continue to eat dolphin meat because they are unaware of the mercury content and dolphin meat is marketed as whale meat.
16.The number of fish are declining due to over fishing. IWC blame dolphins and whales for the declining rate.
18. The form of propaganda to "sell" dolphin meat to people by selling it to schools to tell people that it is healthy and safe.
20. The main character goes to IWC conference room carrying monitors to display the reality of the dolphins.
22. Whaling was banned in 1986
Saturday, June 6, 2015
Monday, June 1, 2015
Photosynthesis vs. Cellular Respiration
Photosynthesis: process where plants and other organisms use sunlight to synthesize food from carbon dioxide and water
Light Dependent Reaction and Light Independent Reaction (Calvin Cycle)
Cellular Respiration: process to produce energy in the form of adenosine triphosphate (ATP) through oxidation of food molecules
1. Glycolysis (Does not require O2)
2. Pyruvate Oxidation
3. Kreb's Cycle
4. Electron Transport Chain/ Chemiosmosis
Light Dependent Reaction and Light Independent Reaction (Calvin Cycle)
Cellular Respiration: process to produce energy in the form of adenosine triphosphate (ATP) through oxidation of food molecules
1. Glycolysis (Does not require O2)
2. Pyruvate Oxidation
3. Kreb's Cycle
4. Electron Transport Chain/ Chemiosmosis
|
|
Cellular Respiration
|
Photosynthesis
|
|
In what
|
Plants, some bacteria, and animals
|
Plants and some bacteria
|
|
Location
|
Glycolysis: Cytoplasm
Krebs Cycle: Mitochondria Matrix
ETC: Inner mitochondrial membrane
|
Chloroplast
|
|
ATP, NADH, FADH2 - Produced
|
Glycolysis: 4 ATP (2 net ATP)
2 NADH
Pyruvate Oxidation: 2 NADH
Kreb's Cycle: 2 ATP
6 NADH
2 FADH2
CO2
Overall: 36-38 ATP
|
18 ATP
12 NADP
|
|
When it can occur
|
When glucose and oxygen is
available
|
When sunlight, water, and
carbon dioxide is available
|
|
Function
|
Break down of food. Energy released
|
Production of food. Energy captured
|
|
Equation
|
6O2 + C6H12O6 ------> 6CO2 +6H2O + ATP (energy)
|
light
6CO2 + 12H2O ---------> C6H12O6 + 6O2 + 6H20 |
Sunday, May 10, 2015
Photosynthesis: Light-dependent reaction vs. Light-independent reaction
Light-dependent reactions: reaction that traps sunlight and produces ATP and NADPH
- Photosystem II is 'excited' because it absorbs a light photon (P680 wavelength) and pulls electrons from H2O (causing hydrogens to break apart from oxygen) and PSII gains electrons. Process of breaking water apart is called photolysis.
Oxidation: element gains an electron
Reduction: element loses an electron
- electrons transferred to PQ, then b6-f and forms a bridge to allow hydrogen atoms to enter the leaf, then electrons transfer to PC
- PSI gets 'excited' with light photon (P700 wavelength) and then becomes oxidized, then electrons pass through Fd, FNA, and NADP
- Once NADP becomes oxidized, hydrogen pair (H2) breaks apart and bonds with NADP+ and becomes NADPH
- ADP synthase bonds with Adenine and 2 phosphate molecules to form ADP; they spin quickly allowing excess hydrogen to exit the leaf.
Then 3rd phosphate atom bonds with ADP, forming ATP (process called Chemiosmosis)

Light-independent reaction: reaction that assimilates CO2 to produce an organic molecule
- carbon dioxide fixation - carbon dioxide chemically bonds with RuBP (ribulose 1,5-biphosphate) breaks into 2 stable PGA's (3-phosphoglycerate). Ribisco helps catalyze this reaction
CO2 + RuBP -> unstable C6 -> PGA
- PGA's are activated by ATP (add phosphate group) and then reduced by NADPH (removes phosphate group). NADPH loses its hydrogen. New compound formed, G3P. G3P may be used to make glucose and carbohydrates.
- reduced G3P make more RuBP - energy (ATP) is required to break and reform the bonds to make 5 carbon RuBP from G3P
- Calvin cycle must be completed 6 times to synthesize 1 molecule of glucose
- 12 G3P that are produced in the 6 cycles, 10 are used to generate RuBP, and 2 are used to make 1 glucose.
6CO2 + 18 ATP + 12 NADPH + water -> 2 G3P + 16 Pi + 18 ADP + 12 NADP+
- Photosystem II is 'excited' because it absorbs a light photon (P680 wavelength) and pulls electrons from H2O (causing hydrogens to break apart from oxygen) and PSII gains electrons. Process of breaking water apart is called photolysis.
Oxidation: element gains an electron
Reduction: element loses an electron
- electrons transferred to PQ, then b6-f and forms a bridge to allow hydrogen atoms to enter the leaf, then electrons transfer to PC
- PSI gets 'excited' with light photon (P700 wavelength) and then becomes oxidized, then electrons pass through Fd, FNA, and NADP
- Once NADP becomes oxidized, hydrogen pair (H2) breaks apart and bonds with NADP+ and becomes NADPH
- ADP synthase bonds with Adenine and 2 phosphate molecules to form ADP; they spin quickly allowing excess hydrogen to exit the leaf.
Then 3rd phosphate atom bonds with ADP, forming ATP (process called Chemiosmosis)

Light-independent reaction: reaction that assimilates CO2 to produce an organic molecule
- carbon dioxide fixation - carbon dioxide chemically bonds with RuBP (ribulose 1,5-biphosphate) breaks into 2 stable PGA's (3-phosphoglycerate). Ribisco helps catalyze this reaction
CO2 + RuBP -> unstable C6 -> PGA
- PGA's are activated by ATP (add phosphate group) and then reduced by NADPH (removes phosphate group). NADPH loses its hydrogen. New compound formed, G3P. G3P may be used to make glucose and carbohydrates.
- reduced G3P make more RuBP - energy (ATP) is required to break and reform the bonds to make 5 carbon RuBP from G3P
- Calvin cycle must be completed 6 times to synthesize 1 molecule of glucose
- 12 G3P that are produced in the 6 cycles, 10 are used to generate RuBP, and 2 are used to make 1 glucose.
6CO2 + 18 ATP + 12 NADPH + water -> 2 G3P + 16 Pi + 18 ADP + 12 NADP+
Sunday, May 3, 2015
Pig Dissection
On April 27 & 28, our grade 12 biology class performed a fetal pig dissection. This dissection was a hands- on opportunity for students to further understand the various systems working in our body; specifically urinary, endocrine, and nervous system.
Intestines: digest & absorb food Stomach: digests & absorbs food;
inside: bile,amniotic fluid, etc.
Spleen: blood cells produced/formed Kidneys: reabsorb water and nutrients, produce urine, filter toxins
Testicles: produce sperm and testosterone Abdominal Cavity
Lungs: bring oxygen into blood and remove carbon dioxide
Trachea: tube to transfer air Heart: pumps blood through circulatory system
Skull: bone to protect brain
Brain: motor control, emotional response, memory, speech, senses, etc.
When the brain is removed Eye
Eye Pupil
Monday, April 6, 2015
Nervous System
Textbook
page 351-353
Structure
of neurons
- has
cell membrane, cytoplasm, mitochondria, nucleus
-
transmits nerve impulses
- different
neurons = different shapes and sizes
4 common features = dendrites, cell
body, axon, branching ends
Dendrites
- receive nerve impulses from neurons/sensory receptors and passes the impulse
to cell body
Cell
Body - contains nucleus; site of metabolic reactions; processes input of
dendrites (if large enough, passes to axon)
Axon
- conducts impulse away from body
Myelin sheath = insulating layer
around axon of some neurons
protects neurons and speeds rate
of nerve impulse transmission
Schwann cells = type of glial cell,
form myelin by wrapping around axon
Classifying
Neurons
1.
Structure - 3 types: multipolar,
bipolar, unipolar neurons
2.
Function -3 types: sensory neurons,
interneurons, motor neurons
The
Reflect Arc
Reflexes - involuntary responses to certain stimuli
Reflex arcs - connections of neurons explaining reflexive
behaviour
- 3 neurons to transmit messages = fast reflexes
(from brain to spinal cord)
1. stimulus (cactus needle) - receptors in skin sense
pressure
2. initiates impulse in sensory neuron
3. activates interneuron in spinal cord
4. signals motor neuron to withdraw hand
Sunday, March 29, 2015
Biotechnology DNA Cloning
Sunday, March 1, 2015
Translation
1) Initiation
2) Elongation
3) Termination
- mRNA transferred to the ribosomes
- Initiation factors (protein) assemble
- Small ribosomal sub-unit - attaches to mRNA near start codon (AUG)
- Initiator tRNA - first to bind with codon (carrying methionine amino acid)
- mRNA
- Large ribosomal sub-unit - to form active ribosome
- 3 binding sites for tRNA
- P (peptide) - contains tRNA with polypeptide attached
- A (amino acid) - contains tRNA with next amino acid added to polypeptide chain
- E (exit) - uncharged tRNA with no amino acid attached exits
- Moves from 5' to 3'
2) Elongation
- tRNA anticodons bind to mRNA codons in the 'A' site of the ribosome
- Initiating transfer RNA binds to the 'P' site
- tRNA recognizes the next codon and second amino acid moves to 'A' site
- Amino acid from tRNA joins amino acid by the tRNA that just entered 'A' site
- First transfer RNA is released
- Next transfer RNA moves to 'A' site and ribosome moves down one codon
- Second amino acid on tRNA in 'P' site transfers to third amino acid
- And Continues: Ribosome to move along mRNA and new amino acids are added to the growing polypeptide chain
3) Termination
- Terminates when mRNA reaches stop codon
- Stop Codons: UAA, UAG, UGA
- Release factor (protein) cuts the polypeptide from last tRNA
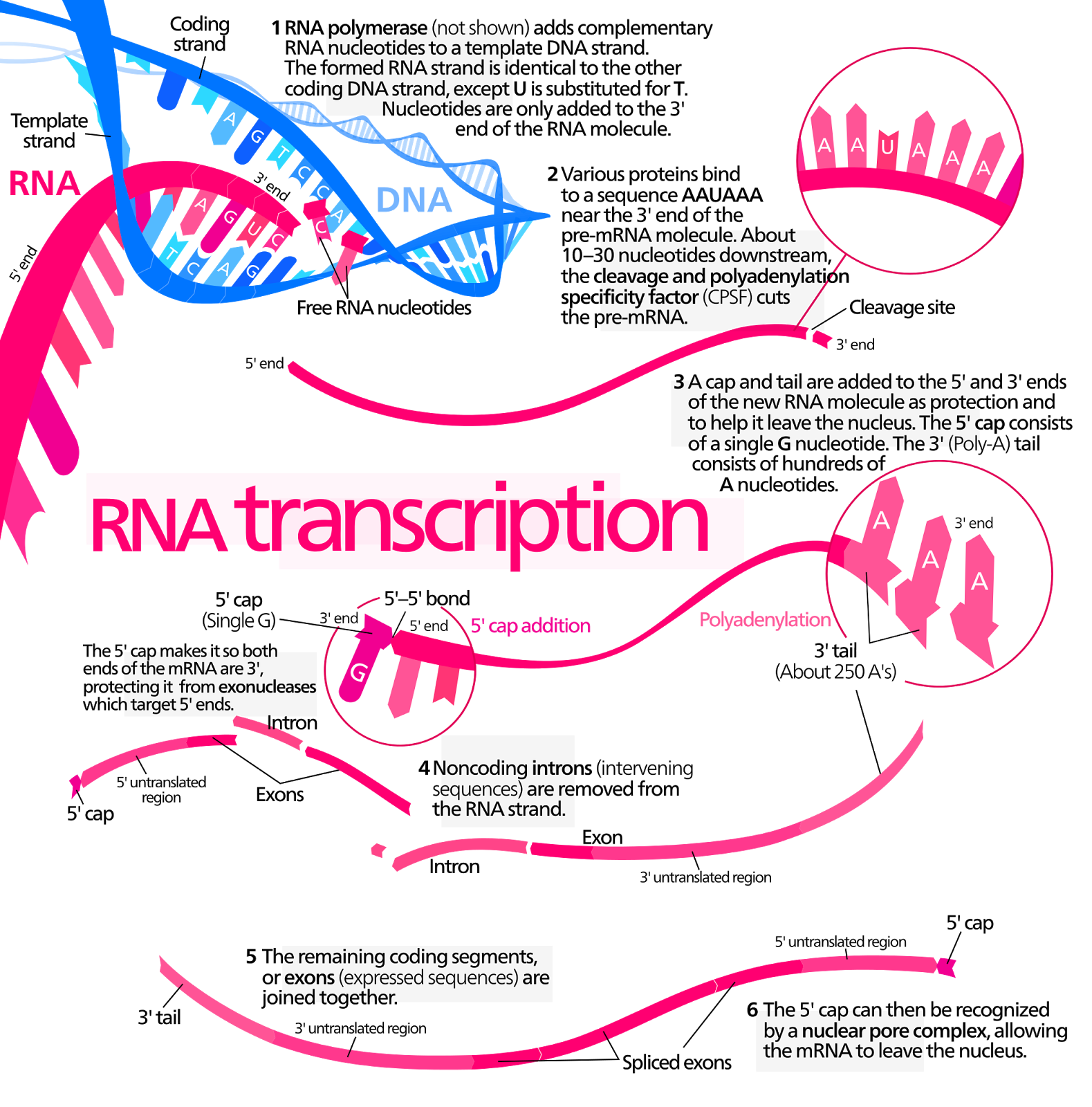
Transcription
1) Initiation
2) Elongation
3) Termination
- Transcription Factors assemble on the promoter region (TATA box)
- RNA polymerase II binds to the transcription factors (transcription initiation complex)
2) Elongation
- Moving from 5' to 3', RNA polymerase synthesizes a strand of pre-mRNA that is complimentary to the template strand of DNA
- Template strand (antisense strand)- the sequence of DNA that is copied
- Coding strand (sense strand)- the sequence that corresponds with
- Pre-mRNA strand, Thymine is replaced with Uracil
3) Termination
- RNA polymerase II reaches the terminator sequence (AAUAA), transcription stops
- Pre-mRNA is removed
- Pre-mRNA protected by G cap (guanine) on the 5' side and poly A tail on the 3' side
- RNA splicing
- Introns (non-coding regions) - are removed from pre-mRNA
- Exons (coding regions) - join to form the mature mRNA
- snRNA + snRNP(proteins) - recognizes regions where exons and introns meet
- snRNP interact with other proteins = larger spliceosome complex that removes introns
- mRNA strand is complete and ready to transport to the ribosomes (factory)
Sunday, February 22, 2015
DNA Replication
The Process of DNA Replication:
1) Initiation
2) Elongation
3) Termination
Note:
- semi-conservative - one parent strand & one daughter strand in the replicated DNA
- new DNA strand can only elongate in the 5' --> 3' direction
1) Initiation
- Helicase - unwinds the double stranded DNA helix
- Single-Strand Binding Proteins - react with single stranded DNA to stabilize it and keep the DNA strands separated
- Gyrase - relieves tension in the DNA from unwinding process
- Primase (RNA Polymerase)
- creates RNA primers (~10 primers; according to parent strand)
- initiates Polymerase III to form complementary strand
2) Elongation
- DNA Polymerase III - attaches to a primer and adds nucleotides to 3' end of the new DNA strand
- 2 types of strands in each strand:
- Leading Strand - from 5' --> 3' into replication fork
- Lagging Strand - from 5' --> 3' away from replication fork; replicates in short segments (Okazaki Fragments) - DNA Polymerase I - replaces Polymerase III when it reaches the primer and replaces the RNA primer with correct DNA sequence
3) Termination
- DNA ligase
- attaches and forms phosphodiester bonds
- connects Okazaki Fragments
Note:
- semi-conservative - one parent strand & one daughter strand in the replicated DNA
- new DNA strand can only elongate in the 5' --> 3' direction
Subscribe to:
Comments (Atom)